Can we predict protein structure directly from sequence w/o MSAs and w/o intermediate steps like distograms? In a new preprint we show that we can, led by
@mrprotein24
@NazimBouatta
@SurgeBiswas
along w/
@charochereau
@geochurch
& Peter Sorger: (1/4)
7
95
374
Replies
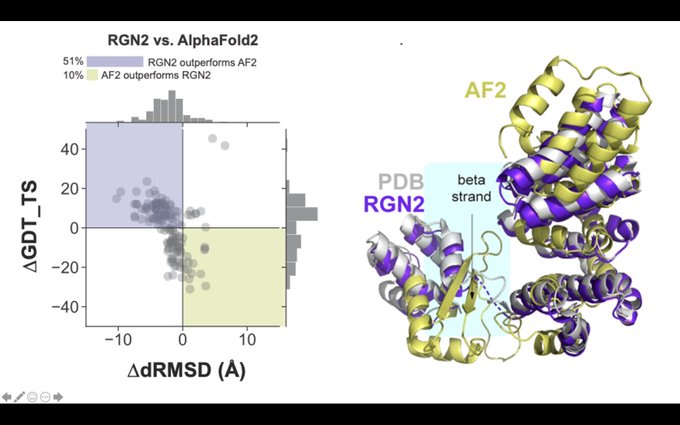
We combine a new protein language model (AminoBERT) with an improved version of our end-to-end differentiable machinery (RGN2) to directly generate 3D coordinates. On orphan proteins, RGN2 outperforms all major methods, including
#AlphaFold
, RoseTTAFold, and trRosetta. (2/4)
2
11
64
On designed proteins RGN2 is close but not yet best accuracy-wise. However, it is orders of magnitude faster; a useful property for exploring new protein sequences. (3/4)
1
2
28
Comments on manuscript are most welcome of course. For future work, we look to combine ideas from AF2 with language models, without sacrificing speed. (4/4)
3
1
17
@MoAlQuraishi
@mrprotein24
@NazimBouatta
@SurgeBiswas
@charochereau
@geochurch
Is there code/model parameters available?
1
0
1
@ThomasJAGraham
@mrprotein24
@NazimBouatta
@SurgeBiswas
@charochereau
@geochurch
Not yet, but will soon!
2
1
11
@MoAlQuraishi
@mrprotein24
@NazimBouatta
@SurgeBiswas
@charochereau
@geochurch
Do you find that weight decay / dropout helps on the pretraining? Most models transformers I see don't use those.
1
0
3
@MoAlQuraishi
@mrprotein24
@NazimBouatta
@SurgeBiswas
@charochereau
@geochurch
Very interesting.
(And very interesting too that this is the second article on this topic this week - the other one by Burkard Rost and col)
1
1
3
@MoAlQuraishi
@mrprotein24
@NazimBouatta
@SurgeBiswas
@charochereau
@geochurch
Is the code available? I am interested in the representations learned by the model.
1
0
2