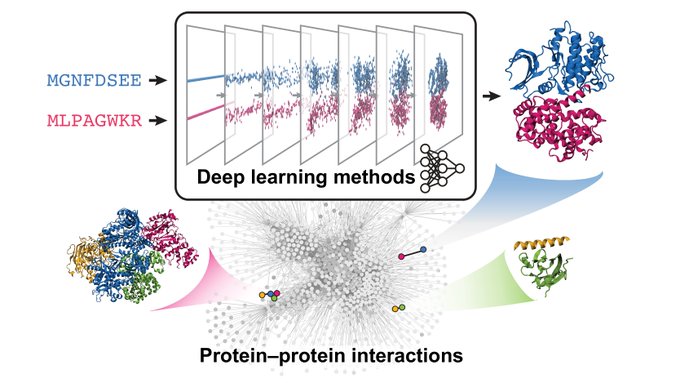
Can we characterize the full diversity of protein interactions that coordinate cell function? Deep learning is a promising way!
@MoAlQuraishi
, Gergő Nikolényi, and I review the ecosystem of DL models for protein interaction discovery, elucidation & design
4
66
321
Replies
DL methods can leverage both heterogeneous data and basic biophysical knowledge to model diverse molecular recognition mechanisms. We discuss key choices encountered across all aspects of model development and their respective trade-offs.
1
0
4
Exciting successes include the use of representation learning to capture complex features pertinent to predicting protein interactions and interaction sites, geometric deep learning to reason over protein structures & generative modeling to de novo design complexes!
1
0
6
Despite these remarkable successes, challenges remain & we outline some promising ideas across all aspects of model development for tackling them. These are truly exciting times to be developing DL models of protein interactions! 4/4
0
0
5