New! We’ve just put up a note evaluating the latest, in-development version of AlphaFold (“AlphaFold-latest”). This is a preview - development is still in progress - but performance across a wide range of tasks is striking.
Highlights in the thread.
1/7
9
270
841
Replies
AlphaFold-latest improves upon AlphaFold 2.3 (the update from late 2022, already a very strong baseline!) for protein-protein structure prediction, especially in hard categories such as bound antibody structures.
2/7
1
10
46
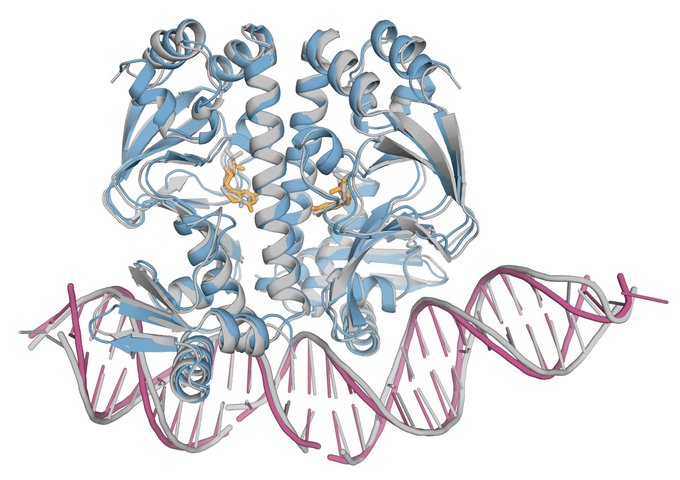
Protein-nucleic complexes make up some of the most important systems like the ribosome. For protein-DNA interfaces AlphaFold-latest outperforms competing systems, while for RNA structure prediction it appears to be competitive with other methods, but more work to do!
3/7
3
9
50
Ligand docking is a key component in comp drug discovery. AF-latest outperforms classical systems like AutoDock Vina on the PoseBusters benchmark. This is despite baselines having access to the ground-truth protein structure information that AlphaFold-latest doesn’t get.
4/7
1
17
73
Finally, many interesting biological processes involve residue modifications, such as glycosylation in proteins. AF-latest can predict the structure of the range of features seen in biomolecules like covalently bound ligands, glycosylation, and modified residues.
5/7
2
8
37
In conclusion, we hope this demonstrates the potential for atomically-accurate structure prediction for the full range of important biomolecules and their interactions using AlphaFold-like methods!
6/7
1
3
32
And here’s my colleague
@maxjaderberg
at
@IsomorphicLabs
’s take on it:
7/7
Excited to show a glimpse of the next generation of AlphaFold from our teams at
@IsomorphicLabs
and
@GoogleDeepMind
. This model expands beyond proteins to include other molecules like small molecules and nucleic acids, and improves accuracy on proteins 1/n
11
143
596
0
4
26
@tfgg2
@tfgg2
Truly groundbreaking stuff. Are you able to comment on when the models will be publicly available?
0
0
17
@tfgg2
Awesome, including post translation modification in protein structure or complex interaction would be huge. How long can we expect to play with it ?
0
0
2
@tfgg2
Great to see these latest developments! Will AlphaFold-latest also be able to include membranes in the prediction? So no interaction possible between internal vs external membrane domains?
0
0
2
@tfgg2
Very interesting, hopefully we will be able to use it on our own projects soon 🤞 also eager to see a comparison to
#RFAA
. 📊
0
0
1